Fusarium
Most Fusarium species are soil fungi and have a worldwide distribution. Some are plant pathogens, causing root and stem rot, vascular wilt or fruit rot.
Several species have emerged as important opportunistic pathogens in humans causing hyalohyphomycosis (especially in burn victims and bone marrow transplant patients), mycotic keratitis and onychomycosis (Guarro 2013). Other species cause storage rot and are important mycotoxin producers.
Multi-locus sequence analysis of EF-1α, β-tubulin, calmodulin, and RPB2 have revealed the presence of multiple cryptic species within each “morphospecies” of medically important fusaria (Balajee et al. 2009). For instance, Fusarium solani represents a complex (i.e. F. solani complex) of over 45 phylogenetically distinct species of which at least 20 are associated with human infections. Similarly, members of the Fusarium oxysporum complex are phylogenetically diverse, as are members of the Fusarium incarnatum-equiseti complex and Fusarium chlamydosporum complex (Balajee et al. 2009, Tortorano et al. 2014, Salah et al. 2015).
Currently the genus Fusarium comprises at least 300 phylogenetically distinct species, 20 species complexes and nine monotypic lineages (Balajee et al. 2009, O’Donnell et al. 2015). Most of the identified opportunistic Fusarium pathogens belong to the F. solani complex, F. oxysporum complex and F. fujikuroi complex. Less frequently encountered are members of the F. incarnatum-equiseti, F. dimerum and F. chlamydosporum complexes, or species such as F. sporotrichioides (O’Donnell et al. 2015, van Diepeningen et al. 2015).
-
Laboratory identification

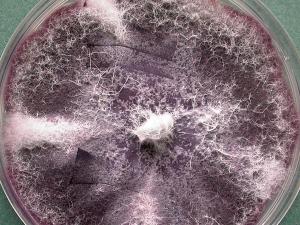
Cultures of F. oxysporum showing purple pigmentation and F. subglutinans showing pink pigmentation.
Morphological description:
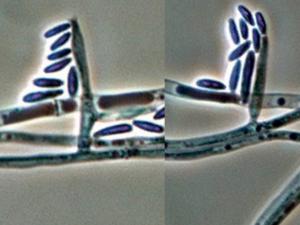
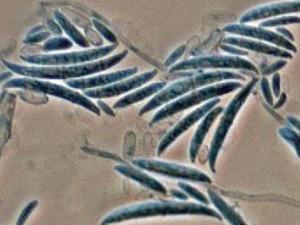
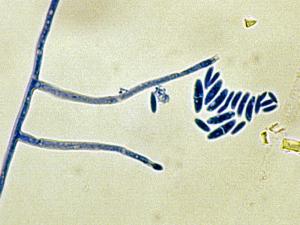
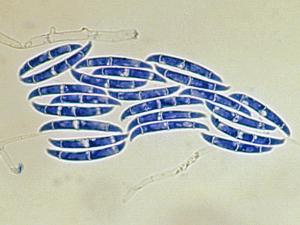
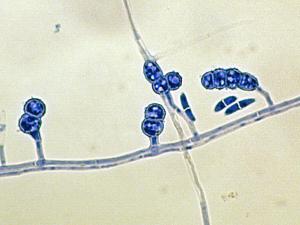
Colonies are usually fast growing, pale or bright-coloured (depending on the species) with or without a cottony aerial mycelium. The colour of the thallus varies from whitish to yellow, pink, red or purple shades. Species of Fusarium typically produce both macro- and microconidia from slender phialides. Macroconidia are hyaline, two to several-celled, fusiform to sickle-shaped, mostly with an elongated apical cell and pedicellate basal cell. Microconidia are one or two-celled, hyaline, smaller than macroconidia, pyriform, fusiform to ovoid, straight or curved. Chlamydospores may be present or absent.Identification of Fusarium species is often difficult due to the variability between isolates (e.g. in shape and size of conidia and colony colour) and because not all features required are always well developed (e.g. the absence of macroconidia in some isolates after subculture).
Note:
Sporulation may need to be induced in some isolates and a good slide culture is essential. The important characters used in the identification of Fusarium species are as follows.- Colony growth diameters on potato dextrose agar and/or potato sucrose agar after incubation in the dark for four days at 25C.
- Culture pigmentation on potato dextrose agar and/or potato sucrose agar after incubation for 10-14 days with daily exposure to light.
- Microscopic morphology including shape of the macroconidia; presence or absence of microconidia; shape and mode of formation of microconidia; nature of the conidiogenous cell bearing microconidia; and presence or absence of chlamydospores.
Molecular identification:
Current species identification is on the basis of multilocus sequence data (Guarro 2013, O’Donnell et al. 2015, van Diepeningen et al. 2015). FUSARIOID-ID (https://www.fusarium.org) is an internet-accessible validated database dedicated to the identification of fusaria via ITS or multi-locus nucleotide BLAST queries, available through the Westerdijk Fungal Biodiversity Institute.For sequence-based identification of Fusarium species (O’Donnell et al. 2015).
- FUSARIOID-ID is the recommended sequence database, rather than GenBank.
- Eight loci may be used, TEF-1, RBP1, RBP2, ACT, Cal, Histone H3, IGS, ITS, 28S, β-tubulin. Use of at least two independent loci will increase the accuracy of identification.
- Ensure sequences are carefully edited and free of ambiguities.
- Ensure the species names associated with the top BLASTn matches are the same. If multiple species names have similar scores it may be necessary to sequence additional loci.
Note:
ITS and D1/D2 sequences are too conserved to resolve species limits of most fusaria. O’Donnell et al. (2015) recommend avoiding ITS or D1/D2 sequences from an unknown isolate to query GenBank, because >50% of the sequences from Fusarium species are misidentified in this database.Identifications based on morphology and/or ITS and D1/D2 sequences should be reported as species complexes. Sequencing of EF-1α, RPB1 and/or RPB2 is required for accurate species identification.
MALDI-TOF MS:
A comprehensive ‘in-house’ database of reference spectra allows accurate identification of Fusarium species complexes (Lau et al. 2013).References:
Booth (1971, 1977), Domsch et al. (1980), McGinnis (1980), Burgess and Liddell (1983), Rippon (1988), Samson et al. (1995), de Hoog et al. (2000, 2015), O’Donnell et al. (2008, 2009a, 2009b, 2015), Balajee et al. (2009), Guarro (2013), Geiser et al. (2013), van Diepeningen et al. (2015), Salah et al. (2015), Tortorano et al. (2014).
Species descriptions
-
Fusarium chlamydosporum complex
Fusarium chlamydosporum complex contains five phylogenetically distinct species and is common in soils and the rhizosphere of numerous vascular plants worldwide. It is occasionally isolated from human and animal infections (O’Donnell et al. 2009b, Guarro 2013).

Fusarium chlamydosporum complex, culture showing pink to ochraceous to brownish surface and a carmine red reverse.
RG-1 organisms.
Morphological description:
Colonies growing rapidly, with abundant aerial mycelium, deep pink, red or ochraceous to brownish; reverse carmine red or tan to brown. Sporodochia orange, flesh-coloured or ochraceous. Conidiophores scattered over the aerial mycelium, branched; numerous polyblastic conidiogenous cells are present. Macroconidia rarely produced and appearing only on sporodochial phialides, usually three-(some up to five)-septate, slightly curved, 30-38 x 3.0-4.5 μm, with no distinct foot-shaped cell. Microconidia and blastoconidia fusiform, rounded apically and tapered towards the base, single-celled to one-(some up to three)-septate, 6-26 x 2-4 μm. Chlamydospores abundant, intercalary, often roughenedAntifungal susceptibility: Fusarium chlamydosporum complex (Australian national data); MIC µg/mL. No ≤0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 ≥32 AmB 11 1 1 4 2 2 1 ISAV 2 2 VORI 11 1 1 7 2 POSA 10 2 5 3 ITRA 11 1 1 2 1 1 5 -
Fusarium dimerum complex
The Fusarium dimerum complex contains 12 phylogenetically distinct species including F. delphinoides, F. penzigii and F. dimerum. These are regarded as cosmopolitan saprotrophs in soil and on plant materials (Domsch et al. 2007). They have also been isolated from human corneal ulcers after trauma and from disseminated or localised infections in immunocompromised patients (Schroers et al. 2009, Guarro 2013).

Fusarium dimerum complex culture showing orange to deep apricot colour due to confluent conidial slime, and macroconidia.
RG-1 organisms.
Morphological description:
Colonies growing slowly; surface usually orange to deep apricot due to confluent conidial slime; aerial mycelium sometimes floccose and whitish. Conidiophores loosely branched, with short, often swollen phialides, 10-18 x 4-5 μm. Macroconidia strongly curved and pointed at the apex, mostly one-(some up to three)-septate, 5-25 (-32) x 1.5-4.2 μm. Microconidia absent. Chlamydospores mostly intercalary, exceptionally terminal, spherical to ovoidal, 6-12 μm diam, smooth-walled, single or in chains.Antifungal susceptibility: Fusarium dimerum complex (Australian national data); MIC µg/mL No ≤0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 ≥32 AmB 42 1 7 10 11 13 ISAV 8 1 3 4 VORI 41 6 12 15 6 2 POSA 39 1 5 8 25 ITRA 42 2 2 38 -
Fusarium fujikuroi complex
Synonymy:
Gibberella fujikuroi complex.Fusarium fujikuroi complex consists of 50 phylogenetically distinct species including 13 of which have been reported to cause human infection; F. acutatum, F. ananatum, F. andiyazi, F. fujikuroi, F. guttiforme, F. napiforme, F. nygamai, F. verticillioides, F. proliferatum, F. sacchari, F. subglutinans, F. temperatum and F. thapsinum (Guarro, 2013, Al-Hatmi et al. 2015).
RG-1 organisms.
Morphological description:
Colonies growing rapidly, pink or vinaceous to violet; aerial mycelium abundant. Sporodochia present or absent, when present they are tan to orange. Conidiophores usually erect and branched. Macroconidia abundant, falcate to rather straight, three to five-septate, with a distinct foot-cell, 27-73 x 3.4-5.2 μm. Blastoconidia straight or slightly curved, two to three-septate, fusiform to lanceolate, with a somewhat pointed, often slightly asymmetrical apical cell and a truncate basal cell, 16-43 x 3.0-4.5 μm. Microconidia produced on polyphialides and aggregating in heads, usually unicellular, ovoidal, ellipsoidal or allantoid, 4-20 x 1.5-4.5 μm. Chlamydospores absent.Antifungal susceptibility: Fusarium fujikuroi complex (Castanheir et al. 2012, Australian national data); MIC µg/mL. No ≤0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 ≥32 AmB 49 2 23 21 2 ISAV 5 1 4 VORI 48 1 17 18 12 POSA 48 4 11 11 9 3 10 ITRA 48 2 3 5 3 35 -
Fusarium incarnatum-equiseti complex
Fusarium incarnatum-equiseti complex consists of 40 phylogenetically distinct species. They occasionally cause infections in humans and animals (O’Donnell et al. 2009b, Guarro 2013).
RG-1 organisms.
Morphological description:
Colonies growing rapidly; aerial mycelium floccose, at first whitish, later becoming avellaneous to buff-brown; reverse pale, becoming peach-coloured. Conidiophores scattered in the aerial mycelium, loosely branched; polyblastic conidiogenous cells abundant. Sporodochial macroconidia slightly curved, with foot-cell, three to seven-septate, 20-46 x 3.0-5.5 µm. Conidia on aerial conidiophores (blastoconidia) usually borne singly on scattered denticles, fusiform to falcate, mostly three to five-septate, 7.5-35 x 2.5-4.0 µm. Microconidia sparse or absent. Chlamydospores sparse, spherical, 10-12 µm diameter, becoming brown, intercalary, single or in chains.Antifungal susceptibility: Fusarium incarnatum-equiseti complex (Australian national data); MIC µg/mL. No ≤0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 ≥32 AmB 10 1 2 1 5 1 ISAV 3 1 1 1 VORI 10 1 2 5 2 POSA 9 1 3 5 ITRA 10 1 1 8 -
Fusarium oxysporum complex
This complex contains at least five phylogenetically distinct species and accounts for about 20% of human infections caused by fusaria (Guarro 2013, Tortorano et al. 2014, Salah et al. 2015). All are ubiquitous soil borne pathogens responsible for vascular wilts, rots, and damping-off diseases of a broad range of plants. A number of these fusaria are also clinically important, causing localised or deeply invasive life threatening infections in humans and other animals (O’Donnell et al. 2009a). Mortality in patients who are persistently and severely neutropenic is typically 100% (Nucci and Anaissie, 2007).
RG-2 organisms.
Morphological description:
Colonies growing rapidly, 4.5 cm in four days, aerial mycelium white, becoming purple, with discrete orange sporodochia present in some strains; reverse hyaline to dark blue or dark purple. Conidiophores are short, single, lateral monophialides in the aerial mycelium, later arranged in densely branched clusters. Macroconidia are fusiform, slightly curved, pointed at the tip, mostly three septate, basal cells pedicellate, 23-54 x 3-4.5 µm. Microconidia are abundant, never in chains, mostly non-septate, ellipsoidal to cylindrical, straight or often curved, 5-12 x 2.3-3.5 µm. Chlamydospores are terminal or intercalary, hyaline, smooth or rough walled, 5-13 µm. In contrast to F. solani complex, the phialides are short and mostly non-septate.Click images below to expand:
Antifungal susceptibility: Fusarium oxysporum complex (Australian national data); MIC µg/mL. No ≤0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 ≥32 AmB 74 1 4 6 17 23 9 12 2 ISAV 13 2 11 VORI 70 5 5 15 28 16 1 POSA 61 1 1 8 11 1 39 ITRA 74 3 1 1 69 -
Fusarium solani complex
The Fusarium solani complex contains at least 60 species and accounts for about 50% of human infections caused by fusaria (Guarro 2013, Tortorano et al. 2014, Salah et al. 2015). All are ubiquitous soil borne pathogens responsible for vascular wilts, rots, and damping-off diseases of a broad range of plants. A number of these fusaria, notably F. keratoplasticum, F. petroliphilum, F. lichenicola and F. solani are clinically important, causing localised or deeply invasive life threatening infections in humans and other animals (Guarro 2013, O’Donnell et al. 2008).
RG-2 organisms.
Morphological description:
Colonies growing rapidly, 4.5 cm in four days, aerial mycelium white to cream, becoming bluish-brown when sporodochia are present. Macroconidia are formed after 4-7 days from short multiple branched conidiophores which may form sporodochia. They are three to five-septate (usually three-septate), fusiform, cylindrical, often moderately curved, with an indistinct pedicellate foot cell and a short blunt apical cell, 28-42 x 4-6 µm. Microconidia are usually abundant, cylindrical to oval, one to two-celled and formed from long lateral phialides, 8-16 x 2-4.5 µm. Chlamydospores are hyaline, globose, smooth to rough-walled, borne singly or in pairs on short lateral hyphal branches or intercalary, 6-10 µm.Click images below to expand:
Antifungal susceptibility: Fusarium solani (Australian national data); MIC µg/mL No ≤0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 ≥32 AmB 239 1 2 19 50 103 56 8 ISAV 48 48 VORI 235 8 25 75 126 POSA 223 2 2 2 2 215 ITRA 239 2 2 233 2